Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
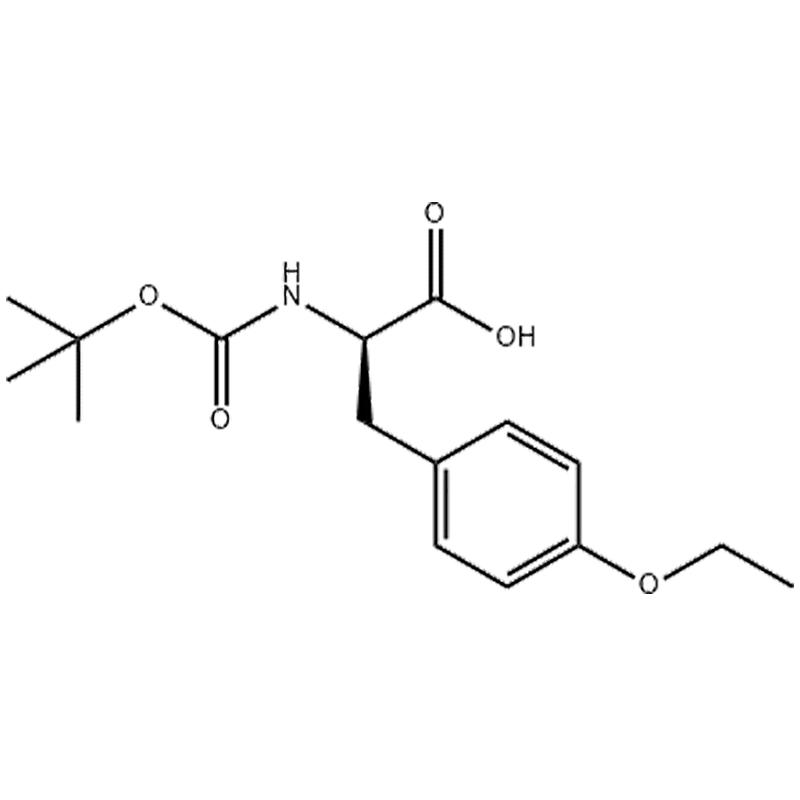
Nature Microbiology (2023 )Cite this article Histidine His

How bacteria link their growth rate to external nutrient conditions is unknown. To investigate how Bacillus subtilis cells alter the rate at which they expand their cell walls as they grow, we compared single-cell growth rates of cells grown under agar pads with the density of moving MreB filaments under a variety of growth conditions. MreB filament density increases proportionally with growth rate. We show that both MreB filament density and growth rate depend on the abundance of Lipid II and murAA, the first gene in the biosynthetic pathway creating the cell wall precursor Lipid II. Lipid II is sensed by the serine/threonine kinase PrkC, which phosphorylates RodZ and other proteins. We show that phosphorylated RodZ increases MreB filament density, which in turn increases cell growth rate. We also show that increasing the activity of this pathway in nutrient-poor media results in cells that elongate faster than wild-type cells, which means that B. subtilis contains spare ‘growth capacity’. We conclude that PrkC functions as a cellular rheostat, enabling fine-tuning of cell growth rates in response to Lipid II in different nutrient conditions.
This is a preview of subscription content, access via your institution
Get full journal access for 1 year
All prices are NET prices. VAT will be added later in the checkout. Tax calculation will be finalised during checkout.
Get time limited or full article access on ReadCube.
All prices are NET prices.
Data generated and analysed during this study are either presented in the paper or in the supplementary materials. Mass proteomic data are available via ProteomeXchange with identifier PXD039295. Source data are provided with this paper.
Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394 (1949).
Kjeldgaard, N. O., Maaloe, O. & Schaechter, M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19, 607–616 (1958).
Article CAS PubMed Google Scholar
Schaechter, M., Maaloe, O. & Kjeldgaard, N. O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19, 592–606 (1958).
Article CAS PubMed Google Scholar
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016).
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Article CAS PubMed PubMed Central Google Scholar
van den Ent, F., Johnson, C. M., Persons, L., de Boer, P. & Lowe, J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29, 1081–1090 (2010).
Article PubMed PubMed Central Google Scholar
Jones, L. J., Carballido-Lopez, R. & Errington, J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001).
Article CAS PubMed Google Scholar
Salje , J. , van den Ent , F. , de Boer , P. & Lowe , J. Direct membrane binding by bacterial actin MreB .Mole.Cell 43, 478–487.
Article CAS PubMed PubMed Central Google Scholar
Hussain, S. et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 7, e32471 (2018).
Dominguez-Escobar, J. et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011).
Article CAS PubMed Google Scholar
Garner, E. C. et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011).
Article CAS PubMed PubMed Central Google Scholar
van Teeffelen, S. et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl Acad. Sci. USA 108, 15822–15827 (2011).
Article PubMed PubMed Central Google Scholar
Dion, M. F. et al. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol. 4, 1294–1305 (2019)..
Billaudeau, C., Yao, Z., Cornilleau, C., Carballido-Lopez, R. & Chastanet, A. MreB forms subdiffraction nanofilaments during active growth in Bacillus subtilis. mBio 10, e01879–18 (2019).
Billaudeau, C. et al. Contrasting mechanisms of growth in two model rod-shaped bacteria. Nat. Commun. 8, 15370 (2017).
Article CAS PubMed PubMed Central Google Scholar
Kock, H., Gerth, U. & Hecker, M. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 51, 1087–1102 (2004).
Article CAS PubMed Google Scholar
Schirner, K. et al. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat. Chem. Biol. 11, 38–45 (2015).
Article CAS PubMed Google Scholar
Meeske, A. J. et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl Acad. Sci. USA 112, 6437–6442 (2015).
Article CAS PubMed PubMed Central Google Scholar
Qiao, Y. et al. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681 (2014).
Article CAS PubMed PubMed Central Google Scholar
Shah, I. M., Laaberki, M. H., Popham, D. L. & Dworkin, J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 (2008).
Article CAS PubMed PubMed Central Google Scholar
Hardt, P. et al. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int. J. Med. Microbiol. 307, 1–10 (2017).
Article CAS PubMed Google Scholar
Kaur, P. et al. LipidII interaction with specific residues of Mycobacterium tuberculosis PknB extracytoplasmic domain governs its optimal activation. Nat. Commun. 10, 1231 (2019).
Article CAS PubMed PubMed Central Google Scholar
Mir, M. et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7, e1002182 (2011).
Article CAS PubMed PubMed Central Google Scholar
Squeglia, F. et al. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J. Am. Chem. Soc. 133, 20676–20679 (2011).
Article CAS PubMed Google Scholar
Wamp, S. et al. PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM. eLife 9, e56048 (2020).
Zheng, C. R., Singh, A., Libby, A., Silver, P. A. & Libby, E. A. Modular and single-cell sensors of bacterial Ser/Thr kinase activity. ACS Synth. Biol. 10, 2340–2350 (2021).
Article CAS PubMed PubMed Central Google Scholar
Madec, E., Laszkiewicz, A., Iwanicki, A., Obuchowski, M. & Seror, S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46, 571–586 (2002).
Article CAS PubMed Google Scholar
Ravikumar, V. et al. In-depth analysis of Bacillus subtilis proteome identifies new ORFs and traces the evolutionary history of modified proteins. Sci. Rep. 8, 17246 (2018).
Article PubMed PubMed Central Google Scholar
Ravikumar, V. et al. Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol. Cell. Proteom. 13, 1965–1978 (2014).
Bendezu, F. O., Hale, C. A., Bernhardt, T. G. & de Boer, P. A. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28, 193–204 (2009).
Article CAS PubMed Google Scholar
Bratton, B. P., Shaevitz, J. W., Gitai, Z. & Morgenstein, R. M. MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. Nat. Commun. 9, 2797 (2018).
Article PubMed PubMed Central Google Scholar
Sauls, J. T. et al. Control of Bacillus subtilis replication initiation during physiological transitions and perturbations. mBio 10, e02205–19 (2019).
Sharpe, M. E., Hauser, P. M., Sharpe, R. G. & Errington, J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol. 180, 547–555 (1998).
Article CAS PubMed PubMed Central Google Scholar
Borkowski, O. et al. Translation elicits a growth rate-dependent, genome-wide, differential protein production in Bacillus subtilis. Mol. Syst. Biol. 12, 870 (2016).
Article PubMed PubMed Central Google Scholar
Lee, S., Wu, L. J. & Errington, J. Microfluidic time-lapse analysis and reevaluation of the Bacillus subtilis cell cycle. Microbiologyopen 8, e876 (2019).
Article CAS PubMed PubMed Central Google Scholar
Hill, N. S., Kadoya, R., Chattoraj, D. K. & Levin, P. A. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 8, e1002549 (2012).
Article CAS PubMed PubMed Central Google Scholar
Cuenot, E. et al. The Ser/Thr kinase PrkC participates in cell wall homeostasis and antimicrobial resistance in Clostridium difficile. Infect. Immun. 87, e00005–19 (2019).
Wamp, S. et al. PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM. eLife 9, e56048 (2020).
Libby, E. A., Reuveni, S. & Dworkin, J. Multisite phosphorylation drives phenotypic variation in (p)ppGpp synthetase-dependent antibiotic tolerance. Nat. Commun. 10, 5133 (2019).
Article PubMed PubMed Central Google Scholar
Pereira, S. F., Goss, L. & Dworkin, J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212 (2011).
Article CAS PubMed PubMed Central Google Scholar
Towbin, B. D. et al. Optimality and sub-optimality in a bacterial growth law. Nat. Commun. 8, 14123 (2017).
Cheng, K. K. et al. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat. Commun. 5, 3233 (2014).
Li, S. H. et al. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947 (2018).
Article CAS PubMed PubMed Central Google Scholar
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Article CAS PubMed Google Scholar
Tinevez, J. Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Article CAS PubMed Google Scholar
Balleza, E., Kim, J. M. & Cluzel, P. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018).
Article CAS PubMed Google Scholar
Waters, J. C. & Wittmann, T. Concepts in quantitative fluorescence microscopy. Methods Cell. Biol. 123, 1–18 (2014).
Spahn, C. et al. DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches. Commun. Biol. 5, 688 (2022).
Article PubMed PubMed Central Google Scholar
Si, F. et al. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr. Biol. 29, 1760–1770.e7 (2019).
Article CAS PubMed PubMed Central Google Scholar
Rubino, F. A. et al. Detection of transport intermediates in the Peptidoglycan Flippase MurJ identifies residues essential for conformational cycling. J. Am. Chem. Soc. 142, 5482–5486 (2020).
Article CAS PubMed PubMed Central Google Scholar
We thank R. Carballido-Lopez, C. Billaudeau and A. Chastanet for helping explore the MreB velocity discrepancy; P. Levin, E. Libby, P. Stoddard and A. Florez for helpful advice and discussions; A. Mollo, S. Rowe and D. Kahne’s lab for biotin-d -Lys and S. aureus PBP4; and L. Lavis for JF dyes. This work was funded by National Institutes of Health Grants DP2AI117923-01 to E.G., as well as support from the Volkswagen Foundation. This work was supported by the NSF-Simons Center for Mathematical and Statistical Analysis of Biology at Harvard (award number 1764269) and the Harvard Quantitative Biology Initiative.
Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA, USA
Yingjie Sun, Sylvia Hürlimann & Ethan Garner
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
E.G. and Y.S. conceived the project and wrote the paper. Y.S. and S.H. conducted all experiments.
The authors declare no competing interests.
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
a, Density of directionally moving MreB-mNeonGreen (bYS09) filaments in cells growing in different media determined from TIRF-SIM imaging. b, Velocity of single molecules of MreB-HaloTag (bYS40), HaloTag-Pbp2a (bYS201), and mNeonGreen-MreC (bYS170) are constant across different growth media - MreB-HaloTag and HaloTag-Pbp2a were labeled with 50 pM of HaloTag-JF549 for 30 min. See also Supplementary Movie 2, 3. c, Velocity of fully labeled MreB-mNeonGreen filaments remains mostly constant across growth rates - Cells expressing MreB-mNeonGreen(bYS09) were grown in different media, and MreB velocity determined by single-molecule tracking and MSD analysis. d, Velocity of MreB-mNeonGreen filaments (bYS09) in cells growing in different media determined by TIRF-SIM imaging. e, MreB velocity remains constant during shifts from poor to rich media - MreB-mNeonGreen (bYS09) cells were grown in S750 glycerol for 6 hs, washed in CH, then placed under a pad made of CH media immediately before imaging with TIRF. Plotted is the growth rate and velocity of directionally moving MreB filaments at each time point. f, Comparison of MreB velocity determined by each group analyzing the MreB-HaloTag (bYS40) data in Extended Data Fig. 1b. The MreB velocity was determined by single-molecule tracking and MSD analysis. g, Comparison of velocities determined by each group analyzing the MreB-mNeonGreen (bYS09) data from the growth shift in Fig. 1d. h, Comparison of velocities determined by each group independently growing and imaging cells containing MreB-msfGFP (bMD159) in rich and minimal media. Carballido-Lopez’s group measured the velocity of MreB-msfGFP in LB and S media. We measured the velocity of MreB-msfGFP in LB, CH, and S750 glycerol. While both groups still disagree, as each continues to attain self-consistant velocities across experiments, both groups remain committed to working together to resolve this discrepancy in the near future. i, Comparison of MreB velocity of MreB-mNeonGreen (bYS09) cells harvested at different OD600 values. j, MreB velocity of MreB-mNeonGreen (bYS09) over time with cells in MatTek dishes vs cells on a CH agar pad sandwiched between two coverslips. The MreB velocity of cells in MatTek dishes was fairly constant over 90 minutes, however the MreB velocity of cells on pads between two coverslips decreases after 20–30 minutes. N values are available in Supplementary Table 2. Boxes indicate 25–75 percentiles, whiskers indicate 10–90 percentiles, the midline indicates median, and + indicates the mean. Error bars and envelopes represent standard deviation. P-values were calculated using one-way ANOVA with multiple comparisons.
a, The abundance of Rod complex components in WT (PY79) cells grown in different media. Abundances are taken from the proteomic mass spectrometry datasets published in Dion et al.13. These datasets included two biological replicates. Each protein abundance is normalized to the mean abundance of that protein in WT cells grown in CH media. Note that membrane proteins, like RodA, were not detected in every replicate. Dotted line represents protein abundances in CH. b, The abundance of Mur enzymes in WT (PY79) cells grown in different media. Abundances are taken from the proteomic mass spectrometry datasets published in Dion et al.13. These datasets included two biological replicates. Each protein abundance is normalized to the mean abundance of that protein in WT cells grown in CH media. Dotted line represents protein abundances in CH. c, The relative abundance of MurAA determined by (Left axis) proteomic mass spectrometry in WT (PY79) and Pxyl-murAA (bYS365) cells growing in glycerol at different xylose concentrations, and (Right axis) the relative intensity of WT murAA-mNeonGreen (bYS397) and Pxyl-murAA-mNeonGreen (bYS1087) cells growing in glycerol at different xylose concentrations. Protein abundances are normalized to WT cells grown in S750 glycerol. The proteomic mass spectrometry dataset included 2–3 biological replicates. Note some error bars are too small to resolve. d, Western blot showing accumulation of Lipid II in WT (PY79) cells grown in S750 glycerol in the presence of cefotaxime (0.25 μg/ml). Fosfomycin (100 μg/ml) depletes Lipid II from cells. The experiment was repeated twice with similar results. e, Western blot showing accumulation of Lipid II in WT (PY79) cells grown in S750 glycerol in the presence of moenomycin (2.5 μg/ml). The experiment was repeated twice with similar results. f, Western blot showing Lipid II levels increases as murAA induction levels in Pxyl-murAA (bYS365). The experiment was repeated twice with similar results. g, MreB velocity remains constant with murAA induction. Cells containing xylose-inducible MurAA and MreB-mNeonGreen (bYS497) were grown in S750 glycerol with a range of MurAA inductions. The velocity of MreB filaments was determined by particle tracking. h, mNeonGreen-MreC velocity under different inductions of Pxyl-murAA (bYS499) grown in S750 glycerol. i, Sub-MIC treatment of moenomycin (2.5 μg/ml) and cefotaxime (0.25 μg/ml) in both CH (left) and S750 glycerol (right) can reduce the growth rate of WT (bYS09) cells without reducing MreB filament density. d, Western blot showing accumulation of Lipid II in WT (PY79) cells grown in S750 glycerol in the presence of cefotaxime (0.25 μg/ml). Fosfomycin (100 μg/ml) depletes Lipid II from cells. e, Western blot showing accumulation of Lipid II in WT (PY79) cells grown in S750 glycerol in the presence of moenomycin (2.5 μg/ml). f, Western blot showing Lipid II levels increases as murAA induction level increases in Pxyl-murAA (bYS365). g, MreB velocity remains constant with murAA induction. Cells containing xylose-inducible MurAA and MreB-mNeonGreen (bYS497) were grown in S750 glycerol with a range of MurAA inductions. The velocity of MreB filaments was determined by particle tracking. h, mNeonGreen-MreC velocity during different inductions of Pxyl-murAA (bYS499) grown in S750 glycerol. i, Sub-MIC treatment of moenomycin (2.5 μg/ml) and cefotaxime (0.25 μg/ml) in both CH (left) and S750 glycerol(right) can reduce the growth rate of WT (bYS09) cells without reducing the MreB filament density. Dashed lines represent the mean of WT values. All constructs are at the native locus unless otherwise indicated. N values are available in Supplementary Table 2. Boxes indicate 25–75 percentiles, whiskers indicate 10–90 percentiles, the midline indicates median, and + indicates the mean. Error bars represent standard deviation.
a, The relative abundance of PrpC and PrkC in WT (PY79) cells grown in different media. Abundances are from the mass spectrometry datasets in Dion et al.13. These datasets included two biological replicates. Each protein abundance is normalized to the mean abundance of that protein in WT cells grown in CH. PrkC is not detected in every replicate. Dotted line represents protein abundances in CH. b, Growth rates of ΔprkC (bYS542), ΔprpC (bYS543), ectopic prkC overexpression (bYS539 with 30 mM xylose), and ectopic prpC overexpression (bYS545 with 30 mM xylose) in cells grown in S750 maltose. c, Growth rates of WT (PY79), ΔprkC (bYS542), and ectopic prkC overexpression (bYS539 with 30 mM xylose) in S750 glycerol, S750 maltose, S750 glucose, CH and LB. d, The relative abundance of YrzL, YpiB, and ClpC/P/X protease in WT (PY79) grown in different media. The abundances are from the mass spectrometry datasets in Dion et al.13. These datasets included two biological replicates. Each protein abundance is normalized to the mean abundance of that protein in WT cells grown in CH. Dotted line represents protein abundances in CH. YrzL and YpiB are not detected in every replicate. Dashed lines represent the mean of WT values. All constructs are at the native locus unless otherwise indicated. N values are available in Supplementary Table 2. Boxes indicate 25–75 percentiles, whiskers indicate 10–90 percentiles, the midline indicates median, and + indicates the mean. P-values were calculated using one-way ANOVA with multiple comparisons.
a and b, PhosTag SDS-PAGE was used to quantitate the fractional phosphorylation of HaloTag-RodZ (labeled with JF549 HaloTag during steady state growth). Separated bands were quantitated with a gel imager. a, PhosTag SDS-PAGE gel of HaloTag-RodZ fusions. Top: SDS-PAGE supplemented with PhosTag causes a band shift with HaloTag-RodZ (bYS163), but not in RodZ(S85A) (bYS165) or ΔprkC (bYS646) cells. Bottom: Regular SDS PAGE gel showing no band shift is observed. The experiment was repeated twice with similar results. b, Top: PhosTag SDS-PAGE of HaloTag-RodZ in cells ectopically overexpressing prkC (bYS643, 30 mM xylose), ectopically overexpressing prpC (bYS644, 30 mM xylose), and under different levels of murAA (bYS642) induction. Bottom: regular SDS PAGE gel, showing no second band is observed. The experiment was repeated twice with similar results. c, Spot plating assay of PY79, ΔprkC (bYS542), and ΔprpC (bYS543) cells on LB agar plates containing 25 μg/ml fosfomycin. The ability of the cells to grow on plates was examined after a 12-hour incubation at 37 °C. d-i, Growth of different strains in a shaking plate reader at 37 °C, where the indicated concentrations of antibiotics were added at time 0. Envelopes represent standard deviation. Each condition was repeated at least 3 times. Note the envelopes on some of the curves are too small to resolve. d, Bulk growth of WT (PY79) cells in CH and S750 glycerol with different concentrations of fosfomycin. e-f, Bulk growth of WT(PY79) and ΔprkC (bYS542) cells grown in CH or S750 glycerol the indicated concentrations of fosfomycin. g-i, ΔprkC (bYS542) shows growth defect relative to WT(PY79) after exposure to sub-MIC levels of antibiotics. Bulk growth curves of WT (PY79) and ΔprkC (bYS542) cells in the presence of sub-MIC (g) moenomycin (5 μg/ml), (h) cefotaxime (0.25 and 0.5 μg/ml), and (i) methilinam (2.5 μg/ml). Dashed lines represent the mean of WT values. All constructs are at the native locus unless otherwise indicated. N values are available in Supplementary Table 2. Envelopes represent standard deviation.
a, Nutrient downshifts of WT (PY79) and ΔprkC (bYS542). Cells were assayed as they were shifted from growth in CH to growth in S750 glycerol. b, c, Cell width decreases (b), and cell length increases (c) with increasing growth rate in the growth accelerated mutants. WT (bME07), RodZ(S85E) (bSKH389), overexpressing murAA (bSKH55) with 30 mM xylose, or ectopically overexpressing prkC (bSKH387) with 30 mM xylose. Unless otherwise indicated, all strains were assayed in S750 glycerol. d, Cell volume remains constant in growth accelerated mutants- Volume for each cell is calculated as (π/6) D2+ (π/4) D2 (L-D), where D is the cell width and L is the cell length. e, The interdivision time of cells decreases as the accelerated cells grow faster – Interdivision time was measured by measuring the time between Z ring constrictions in mNeonGreen-pbp2B in S750 glycerol: WT cells (bME07), cells containing RodZ(S85E) (bSKH389), cells overexpressing murAA (bSKH55) with 30 mM xylose, or cells ectopically overexpressing prkC (bSKH387) with 30 mM xylose. Dashed lines represent the mean of WT values. All constructs are at the native locus unless otherwise indicated. N values are available in Supplementary Table 2. Error bars and envelopes represent standard deviation.
Supplementary Discussions 1 and 2, Notes, Tables 1–3 and References.
Imaging of MreB filaments in different media. TIRF-SIM imaging of MreB-mNeonGreen (bYS09) cells grown in different media and imaged with TIRF-SIM microscopy at 1 s intervals using 100 ms exposures.
Single-molecule imaging of MreB-HaloTag sparsely labelled with JF549. MreB-HaloTag (bYS40) cells were grown in different media, incubated with 50 pM of HaloTag-JF549 ligand in that same media for 30 min, then washed and prepared as indicated in methods. Cells were imaged at 0.5 s intervals using TIRFM with 0.5 s exposures.
Imaging of mNeonGreen-MreC in different media. TIRF imaging of cells expressing mNeonGreen-MreC at the native locus (bYS170) grown in different media. Cells were imaged at 1 s intervals using TIRFM with 1 s exposures.
Imaging of MreB filaments during nutrient upshifts. TIRF-SIM imaging of MreB-mNeonGreen (bYS09) during a nutrient upshift from S750 maltose to CH starting at time 0. Cells were imaged at 1 s intervals with TIRF-SIM using 100 ms exposures.
Imaging of MreB filaments during different levels of murAA induction and murAA depletion. TIRF-SIM imaging of (1) MreB-mNeonGreen (bYS09), (2) MreB-mNeonGreen, Pxyl-murAA (bYS497) with different levels of induction indicated at the bottom of the panel and (3) a MurAA depletion. For the MurAA depletion, bYS497 was first grown in CH with 0.5 mM xylose, washed two times with CH, then resuspended in fresh CH media without xylose and grown for 4 h. All cells were imaged with TIRF at 1 s intervals using 300 ms exposures.
TIRF imaging shows no directional motions of single molecules of MraY, MurG, MurJ or Amj. All cells were grown in CH and imaged at 1 s intervals with TIRF using 300 ms exposures. Column 1: imaging of Phyperspank-HaloTag-MurG (bYS993) induced with 100 uM IPTG. Column 2: imaging of Pxyl-mNeonGreen-MraY (bYS453) induced with 1 mM xylose. Column 3: imaging of Pxyl-mNeonGreen-MurJ (bYS609) induced with 1 mM xylose. Column 4: imaging of Pxyl-mNeonGreen-Amj (bYS611) induced with 1 mM xylose.
Imaging of MreB filaments during MurJ and Amj depletions. Cells were grown in CH or S750 glycerol and imaged at 1 s intervals with TIRF using 300 ms exposures. Column 1: MreB-mNeonGreen (bYS09) growing in CH or S750 glycerol. Column 2: MreB-mNeonGreen, Pxyl-MurJ, Pxyl-Amj (bYS670) cells induced with 0.5 mM xylose growing in CH or S750 glycerol.
PrkC-mediated acceleration of cell growth. Phase-contrast time lapses comparing the growth of WT (PY79) cells grown in S750 glycerol to cells growing in the same media where PrkC was ectopically overexpressed (bYS539) with 30 mM xylose. Cells were imaged using phase contrast at 2 min intervals with 300 ms exposures.
TIRF imaging of HaloTag-PrkC and HaloTag-RodZ. The first series shows HaloTag-PrkC (bYS765) growing in CH and S750 glycerol. HaloTag-PrkC (bYS765) was incubated with 1 uM HaloTag-JF549 ligand for 30 min, washed and then prepared as detailed in methods. The second series shows HaloTag-RodZ (bYS163) growing in CH and S750 glycerol. HaloTag-RodZ (bYS163) was incubated with 1 uM HaloTag-JF549 ligand for 30 min, washed, then imaged. Cells were imaged using TIRFM at 1 s intervals with 0.5 s exposures.
TIRF imaging of MreB with RodZ phosphomutants. MreB-mNeonGreen (bYS09), MreB-mNeonGreen with RodZ(S85A) (bYS748) and MreB-mNeonGreen with RodZ(S85E) (bYS749) cells were grown in S750 maltose and S750 glycerol. All cells were imaged at 1 s intervals using TIRFM with 300 ms exposures.
Source data for Extended Data Fig. 1.
Source data for Extended Data Fig. 2.
Source data for Extended Data Fig. 3.
Source data for Extended Data Fig. 4.
Source data for Extended Data Fig. 5.
Unprocessed regular SDS–PAGE, PhosTag gels and western blots gels for lipid II detection.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Sun, Y., Hürlimann, S. & Garner, E. Growth rate is modulated by monitoring cell wall precursors in Bacillus subtilis. Nat Microbiol (2023). https://doi.org/10.1038/s41564-023-01329-7
DOI: https://doi.org/10.1038/s41564-023-01329-7
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Nature Microbiology (Nat Microbiol) ISSN 2058-5276 (online)
Glutamic Acid Glu Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.